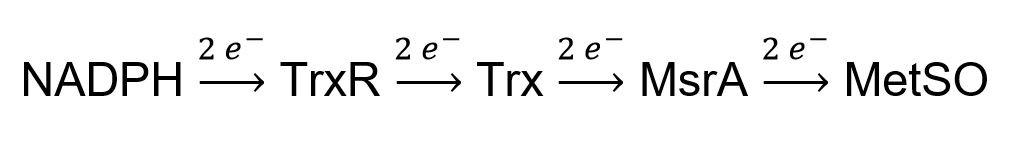Characterization of ancient thioredoxins
Characterization of ancient thioredoxins
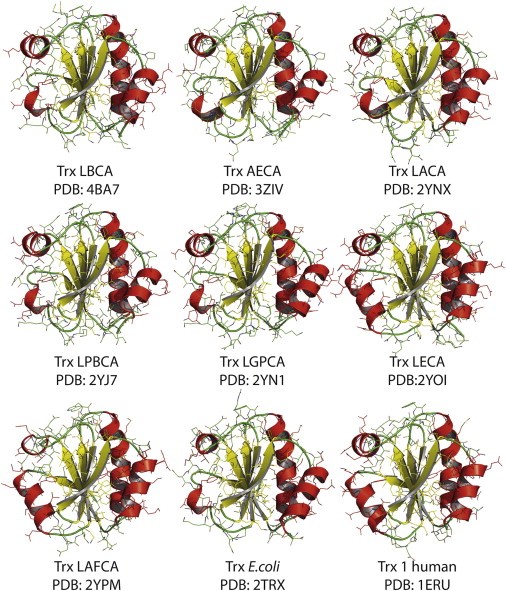
The thiol/disulfide oxidoreductase thioredoxin (Trx) is an ubiquitous, cytosolic enzyme that catalyzes the reduction of numerous protein substrates by NADPH. Specifically, Trx receives two electrons from NADPH via thioredoxin reductase (TrxR) and passes them on to its protein substrates via disulfide exchange. Recently an extensive phylogenetic study was carried out in which Precambrian thioredoxins, dating back between 1.4 and 4 billion years, were resurrected (Structure 21, 1690-1699, 2013). All these Precambrian enzymes displayed the canonical Trx fold and only show small structural changes relative to today’s enzymes. We are performing a biochemical and biophysical characterization of these Precambrian thioredoxins to elucidate whether typical features of today’s thioredoxins, such as their strongly reducing redox potential and high disulfide reductase activity, were already present in ancient enzymes, and whether ancient thioredoxins can still interact efficiently with present-day substrates and thioredoxin reductases in vivo and in vitro. We expect novel insights into the evolution of this ubiquitous protein and its role in the evolution of oxidative stress response.
Specifically, we are addressing the following questions:
- Can ancient thioredoxin still interact with today’s E.coli enzymes, including thioredoxin reductase (TrxR) and methionine sulfoxide reductase (MsrA) and complement thioredoxin deficiency in a trx deletion strain under selective pressure
- Can we reconstitute the entire electron flow pathway from NADPH to methionine sulfoxide (MetSO) from purified components in vitro for quantitative comparison of the reductase activity of ancient thioredoxin with E. coli Trx?
- Is the highly reducing redox potential of the active-site cysteine pair of E. coli thioredoxin preserved throughout the molecular evolution of thioredoxins?
- Is the folding mechanism of ancient thioredoxin similar to that of E. coli thioredoxin where the folding rate is dictated by proline trans→cis isomerization?